Voltammetric Solvation Study for the Interaction of CuBr2 with Oxalic Acid in KBr Solution at 180C Using Glassy Carbon Electrode
Esam.A.Gomaa1*
Adel.Z. EL-Sonbati2
Mostafa.A.Diab3
Mohamed.S.EL-Ghareib4
Hanaa.M.Salama5
1Chemistry Department, Faculty of Science, Mansoura University, Mansoura, Egypt. |
AbstractThe electrochemical behaviour for CuBr2 was studied in 0.1M KBr in absence and presence of different oxalic acid concentrations as ligand. The mechanism for the reduction and oxidation for CuBr2 in absence and presence of oxalic acid ligand were examined. The scan rates were done for the redox reaction of CuBr2 in absence and presence of oxalic acid for explanation of the type for the electrochemical reaction under consideration. Stability constants and complexation Gibbs free energies for the interaction of CuBr2 with oxalic acid were evaluated from the experimental part and their values were discussed. |
Licensed: |
|
Keywords: |
|
Received: 8 July 2020 |
|
| (* Corresponding Author) |
Funding: This study received no specific financial support. |
Competing Interests:The authors declare that they have no competing interests. |
1. Introduction
The heavy metal ions known as pollutants and their interaction by electrochemical methods are interesting [1-6]. Some metal ions can be examined by the reduction at different cathodic materials [6, 7]. In this work, the estimation and evaluation of electrochemical voltammetry energies of CuBr2 in 0.1 M KBr were studied to explain the redox mechanism. The binding of any copper salt with ligands is treated as remediation of copper ions in vitro and vivo [7]. Copper in big quantity cause headache, nose irritation, mouth and eye irritation. It can damage the kidney and liver [7]. Copper is the biggest third abundant metal ions in human body [8]. Excess of copper may cause vomiting and diarrhea [7, 8].
2. Experimental
The chemical CuBr2, KBr are of highly pure from sigma Aldrich Company. Oxalic acid from Adwic company was also used. All chemicals not pre-treated to prevent any harmful effect on them. Conductivity deionized water was used with its conductance value of 2.7 µs. N2 flow was done for 10 minutes to remove the dissolved oxygen for each run made. (G C E) glassy carbon electrode was prepared in our laboratory by joining pure carbon piece with copper wire and covered with heat shrink polymer to isolate it for different ions migration. The glassy carbon electrode (G C E) is polished well before each run with Al2O3on wool piece. The electrode area is 0.502cm2.
3. Results and Discussion
3.1. Cyclic Voltammetry of CuBr2 in Absence of Oxalic Acid
The cyclic voltammetry for the redox behaviour of CuBr2in 0.1M KBr of 180C was studied in range of from 0.6 to -0.8V. The reduction process proceeds in the range 0.6 to -0.8 V and the oxidation in the range -0.8 to 0.6V. Studying the effect of scan rate was done and presented in Figure 1 change of Cu(II) to Cu(O) by the two reduction steps at 0 V and -0.42 V are clear in Figures 1 & 2 .One electron mechanism was suggested for each reduction step. CuBr2shows also two oxidation peaks at -.18V and 0.2V indicating the reverse of the reduction process as explained in next suggested mechanism:
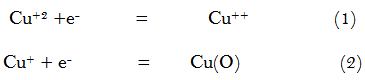
The electrochemical kinetic parameters like electron transfer rate constant ks , cathodic surface coverage (Γc) , anodic surface coverage (Γa) , cathodic quantity of electricity (Qc) , anodic quantity of electricity (Qa), diffusion coefficients for anode and cathode and αna are presented in Table 1 & 2 [8-20]. The data given in Table (1& 2 ) indicates the increase in all kinetic parameters by more adding CuBr2 in solution favouring more diffusion controlled reductions.
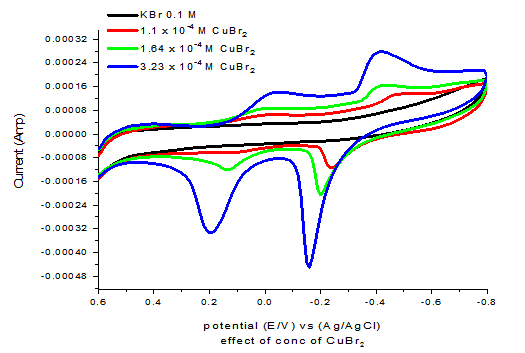
From scan rate effect the different kinetic parameters mentioned before was done and listed in Table 3& 4 explaining also with Figure 2 that the reactions under or redox system is quasi reversible and diffusion controlled reaction.
3.2. Electrochemical Behaviour of Cubr2in Presence of Oxalic Acid as Complexing Agent
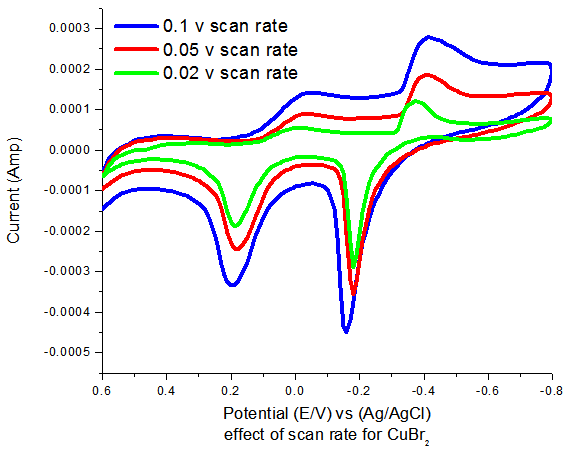
We used oxalic acid as ligand by different concentrations ranging from 9.523x10-4 to 0.0103 M.
By increasing oxalic acid concentration shift in CuBr2 waves specially reduction waves to more negative potentials. Also positive potential shift in oxidation peaks are observed indicating complex behaviour observed and complexation interaction between oxalic acid and copper bromide.
New reduction wave was observed at approximately 0.45V which is suggested as complex of oxalate wave. This wave or peak happens at more positive potentials indicating the ease of its formation.
The stability constant and Gibbs free energies of complexation were calculated and their values represented in Table 5 & 6. These data are obtained from CuBr2 wave analysis for the first reduction wave with the first oxidation peak. Also the second reduction peak with the second oxidation peak.
The obtained thermodynamic parameters evaluated are representing in Table 7 & 8.
The scan rate wave studied and from Figure 4 supported the quasi reversible and diffusion control of redox reaction under consideration.
On drawing the relation between![]() versus scan rate (υ) for complex 1:1 (molar ratio ) we can support the diffusion controlled system of redox reactions.
versus scan rate (υ) for complex 1:1 (molar ratio ) we can support the diffusion controlled system of redox reactions.
For more calculations, the stability complexation constant (Bc ) and the Gibbs energies of complexation (Gc) were evaluated following the redox peak and the data are given in Table 9 & 10.
From the data given in Table 9 &10 we conclude complexation reaction between the salt and oxalic acid.
| Figure-1. Cyclic voltammograms of different concentrations of CuBr2 in 0.1M KBr. |
[M] x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
-ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-8 |
Dc x10-10 |
Epc/2 |
αna |
ks x10-6 |
Γc x10-9 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-5 |
|
0.4 |
1.1 |
-0.23717 |
2.2874E-05 |
-0.23719 |
0.779 |
1.89 |
4.122946874 |
0.564065 |
3.32 |
0.067903901 |
0.688209575 |
0.589 |
1.25577 |
0.951 |
-0.517745 |
-3.92 |
0.6 |
1.64 |
-0.19974 |
-0.001436 |
-0.1983 |
1.49 |
3.83 |
3.887611437 |
0.922958 |
6.11 |
0.075147348 |
0.610006663 |
1.63 |
2.54139 |
1.92 |
-0.987994 |
-7.48 |
1.2 |
3.23 |
-0.15533 |
-0.0403252 |
-0.11501 |
4.26 |
11.6 |
3.680567157 |
1.95657 |
14.4 |
0.059939264 |
0.465931343 |
11.5 |
7.69068 |
5.82 |
-2.83061 |
-21.4 |
ml of Metal |
[M] x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-5 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-9 |
Dc x10-10 |
Epc/2 |
αna |
ks x101 |
Γc x10-9 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-5 |
0.4 |
1.1 |
0.109405 |
-0.4759611 |
0.585366 |
-1.51 |
2.52 |
0.60007111 |
0.211938 |
5.89 |
-0.44102695 |
1.337269069 |
1.38 |
1.67245 |
1.27 |
-0.100359 |
-0.760 |
0.6 |
1.64 |
0.136362 |
-0.4214486 |
0.557811 |
-6.54 |
4.14 |
1.580514509 |
1.78683 |
7.15 |
-0.38207449 |
1.186473391 |
0.827 |
2.75047 |
2.08 |
-0.434716 |
-3.29 |
1.2 |
3.23 |
0.195592 |
-0.4159459 |
0.611537 |
-22.0 |
13.8 |
1.594762346 |
5.19458 |
20.4 |
-0.35701114 |
0.792679151 |
3.32 |
9.14557 |
6.93 |
-1.4585 |
-11.0 |
ʋ |
ν^(1/2) |
ml added from M |
M x10-3 |
-Ep,a (volt) |
-Ep,c (volt) |
∆Ep (volt) |
-ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-10 |
Dc x10-11 |
-Epc/2 |
αna |
ks x10-3 |
Γc x10-8 (mol/cm2) |
(+) Qc x10-4 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
0.05 |
0.223606798 |
1.2 |
1.61 |
0.178058059 |
0.40550807 |
227.450011 |
3.22 |
9.95 |
3.232645273 |
8.92 |
8.53893 |
0.35847049 |
0.993171247 |
2.60 |
1.32226 |
1.00 |
-4.27441 |
-3.24 |
0.02 |
0.141421356 |
1.2 |
1.61 |
0.181402084 |
0.373575153 |
192.173069 |
3.01 |
7.64 |
3.934172239 |
19.5 |
12.5849 |
0.33292042 |
1.149100587 |
1.07 |
2.53812 |
1.92 |
-9.98539 |
-7.56 |
0.01 |
0.1 |
1.2 |
1.61 |
0.145184582 |
0.367994078 |
222.809496 |
4.17 |
11.6 |
3.585229172 |
75.0 |
58.3782 |
0.31935987 |
0.960566209 |
2.73 |
7.73084 |
5.86 |
-27.7168 |
-21.0 |
ʋ |
ν^(1/2) |
ml added from M |
M x10-3 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-10 |
Dc x10-11 |
Epc/2 |
αna |
ks x10-3 |
Γc x10-8 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
0.05 |
0.223606798 |
1.2 |
1.61 |
0.185063824 |
-0.035491563 |
220.5553865 |
-2.07 |
6.75 |
3.06910674 |
3.70 |
3.92454 |
0.053588131 |
0.524433472 |
1.12 |
0.896419 |
6.79 |
-2.7512 |
-2.08 |
0.02 |
0.141421356 |
1.2 |
1.61 |
0.186240013 |
-0.009595429 |
195.8354417 |
-1.68 |
3.32 |
5.070239441 |
6.10 |
2.37409 |
0.063300436 |
0.640864513 |
0.372 |
1.10239 |
8.35 |
-5.58937 |
-4.23 |
0.01 |
0.1 |
1.2 |
1.61 |
0.213753827 |
0.000540333 |
213.2134943 |
-2.04 |
8.94 |
2.282722711 |
18.0 |
34.4686 |
0.091806432 |
0.511869941 |
1.26 |
5.94037 |
45.0 |
-13.5602 |
-10.3 |
ml of Ligand |
[L] x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-8 |
Dc x10-10 |
Epc/2 |
αna |
ks x10-2 |
Γc x10-9 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
|||||||||
0.3 |
0.8 |
-0.25584 |
-0.4776154 |
0.221779 |
-3.68 |
11.5 |
3.195726603 |
1.45769 |
14.3 |
-0.41597888 |
0.757933405 |
1.17 |
7.64529 |
5.79 |
-2.44323 |
-1.85 |
|||||||||
0.6 |
1.59 |
-0.02906 |
-0.2541887 |
0.225124 |
-4.62 |
9.45 |
4.885856274 |
2.29509 |
9.61 |
-0.21310133 |
1.137001723 |
1.26 |
6.27466 |
4.75 |
-3.06571 |
-2.32 |
|||||||||
0.7 |
1.85 |
-0.01884 |
-0.2563617 |
0.237522 |
-4.49 |
8.58 |
5.231597488 |
2.17122 |
7.93 |
-0.20417525 |
0.895182833 |
1.30 |
5.69966 |
4.32 |
-2.98183 |
-2.26 |
|||||||||
0.9 |
2.36 |
-0.03474 |
-0.2944109 |
0.259675 |
-4.62 |
9.67 |
4.780077086 |
2.30072 |
10.1 |
-0.2414744 |
0.882498832 |
2.26 |
6.42139 |
4.86 |
-3.06947 |
-2.32 |
|||||||||
ml of Ligand |
[L] x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-9 |
Dc x10-10 |
Epc/2 |
αna |
ks x10-3 |
Γc x10-9 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
0.3 |
0.8 |
0.102352322 |
-0.129539981 |
0.231892303 |
-2.33 |
7.81 |
2.988446399 |
5.87157 |
6.57 |
-0.01480769 |
0.407177199 |
7.14 |
5.18875 |
3.93 |
-1.55063 |
-1.17 |
0.6 |
1.59 |
0.32043734 |
0.059015852 |
0.261421488 |
-2.25 |
5.17 |
4.351156285 |
5.44593 |
2.88 |
0.167428463 |
0.430912718 |
8.74 |
3.43212 |
2.60 |
-1.49337 |
-1.13 |
0.7 |
1.85 |
0.332309219 |
0.021059568 |
0.311249652 |
-1.96 |
2.92 |
6.723904263 |
4.14532 |
0.917 |
0.102439928 |
0.574049718 |
15.3 |
1.93771 |
1.47 |
-1.3029 |
-0.987 |
0.9 |
2.36 |
0.326947691 |
0.040538909 |
0.286408782 |
-1.93 |
3.56 |
5.403273208 |
3.99763 |
1.37 |
0.152364759 |
0.417760053 |
9.75 |
2.36797 |
1.79 |
-1.27948 |
-0.969 |
Ml of ligand |
[L] X10-4 |
(Ep,a)M |
(Ep,a)C |
∆E mv |
j |
Log βj |
∆G (KJ/mol) |
∆H (KJ/mol) |
∆S (KJ/mol) |
0.3 |
8.00E-05 |
-0.155331539 |
-0.25583682 |
0.100505281 |
0.25 |
4.493734173 |
-25.12001045 |
25.34085784 |
0.172840789 |
0.6 |
1.59E-04 |
-0.155331539 |
-0.029064641 |
0.126266898 |
0.5 |
6.258484448 |
-34.9849788 |
18.19531226 |
0.182155475 |
0.7 |
1.85E-04 |
-0.155331539 |
-0.018839231 |
0.136492308 |
0.583333333 |
6.889700905 |
-38.51348391 |
16.52830513 |
0.18853156 |
0.9 |
2.36E-04 |
-0.155331539 |
-0.034736254 |
0.120595285 |
0.75 |
6.883038331 |
-38.47624007 |
16.54430404 |
0.188458791 |
Ml of ligand |
[L] X10-4 |
(Ep,a)M |
(Ep,a)C |
∆E mv |
j |
Log βj |
∆G (KJ/mol) |
∆H (KJ/mol) |
∆S (KJ/mol) |
0.3 |
8.00E-05 |
0.1955915 |
0.102352322 |
0.093239178 |
0.25 |
-2.194448637 |
12.26698567 |
-51.89234185 |
-0.219761355 |
0.6 |
1.59E-04 |
0.1955915 |
0.32043734 |
0.12484584 |
0.5 |
6.20942863 |
-34.71075638 |
18.33905913 |
0.181708565 |
0.7 |
1.85E-04 |
0.1955915 |
0.332309219 |
0.136717719 |
0.583333333 |
6.897482237 |
-38.55698162 |
16.50965887 |
0.188616683 |
0.9 |
2.36E-04 |
0.1955915 |
0.326947691 |
0.131356191 |
0.75 |
7.254511691 |
-40.55277916 |
15.69713906 |
0.192669698 |
| Figure-2. Effect of different scan rates on cyclic voltamogram of CuBr2. |
| Figure-3. Effect of different concentrations of oxalic acid on the cyclic voltamogram of CuBr2. |
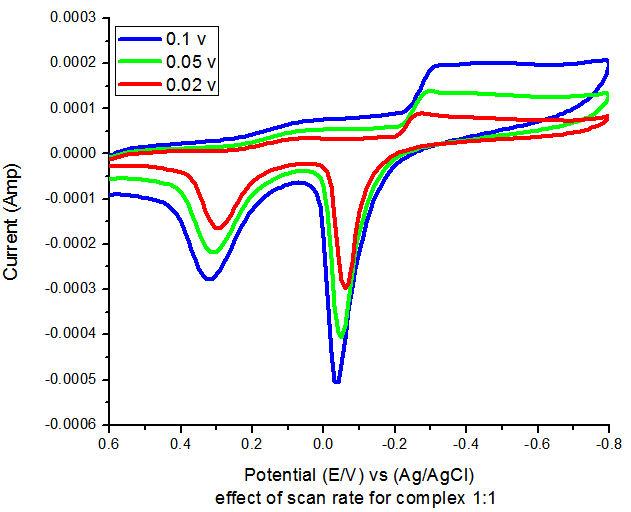
| Figure-4. Effect of different scan rates on the cyclic voltamogram of oxalic acid + CuBr2. |
Effect of different scan rates as seen in Figure 3 indicate the diffusion controlled reaction which decreases by the decrease in san rate vales.
ʋ |
ν^(1/2) |
ml added from M |
M x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-8 |
Dc x10-8 |
Epc/2 |
αna |
ks x10-2 |
Γc x10-8 (mol/cm2) |
(+) Qc x10-4 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
0.05 |
0.223606798 |
1.2 |
3.23 |
-0.048797293 |
-0.295931265 |
247.1339724 |
-3.91 |
8.00 |
4.885503275 |
3.29114 |
1.18933 |
-0.25016663 |
1.183799434 |
1.57 |
1.0627 |
0.805 |
-5.19182 |
-3.93 |
0.02 |
0.141421356 |
1.2 |
3.23 |
-0.064033542 |
-0.267851036 |
203.8174936 |
-2.98 |
4.74 |
6.301518508 |
4.79888 |
0.120851 |
-0.233770273 |
1.370754902 |
0.455 |
1.57305 |
1.19 |
-9.91257 |
-7.51 |
0.01 |
0.1 |
1.2 |
3.23 |
-0.033185921 |
-0.306026067 |
272.8401463 |
-4.97 |
10.5 |
4.728996576 |
26.5975 |
1.18933 |
-0.266562986 |
0.511869941 |
3.69 |
6.97884 |
5.29 |
-33.0029 |
-25.0 |
ʋ |
ν^(1/2) |
ml added from M |
M x10-4 |
Ep,a (volt) |
Ep,c (volt) |
∆Ep (volt) |
ipa x10-4 (Amp) |
ipc x10-5 (Amp) |
-ipa/ ipc |
Da x10-19 |
Dc x10-10 |
Epc/2 |
αna |
ks x10-3 |
Γc x10-9 (mol/cm2) |
(+) Qc x10-5 |
Γa x10-8 (mol/cm2) |
(-)Qa 10-4 |
0.05 |
0.223606798 |
1.2 |
3.23 |
0.308701313 |
0.065304612 |
243.396701 |
-1.50 |
3.15 |
4.750104674 |
4.82573 |
2.13873 |
0.159342368 |
0.496783153 |
4.00 |
4.18528 |
3.17 |
-1.98805 |
-1.51 |
0.02 |
0.141421356 |
1.2 |
3.23 |
0.294298648 |
0.074634865 |
219.6637832 |
-1.13 |
2.42 |
4.656409523 |
6.84191 |
3.15555 |
0.168540324 |
0.497483036 |
1.92 |
8.03811 |
6.09 |
-3.74287 |
-2.83 |
0.01 |
0.1 |
1.2 |
3.23 |
0.327533031 |
0.040381735 |
287.1512965 |
-1.98 |
3.54 |
5.585784971 |
42.1681 |
13.515 |
0.146945123 |
0.43839046 |
10.1 |
23.5255 |
17.8 |
-13.1409 |
-9.95 |
Ml of ligand |
[L] X10-4 |
(Ep,a)M |
(Ep,a)C |
∆E mv |
j |
Log βj |
∆G (KJ/mol) |
∆H (KJ/mol) |
∆S (KJ/mol) |
0.9 |
2.36E-04 |
-0.155331539 |
-0.034736254 |
0.120595285 |
0.75 |
6.883038331 |
-38.47624007 |
76.1531196 |
0.392633532 |
0.9 |
2.36E-04 |
-0.145184582 |
-0.033185921 |
0.111998661 |
0.75 |
6.586277368 |
-36.81734388 |
81.04029714 |
0.403691183 |
0.9 |
2.36E-04 |
-0.181402084 |
-0.064033542 |
0.117368542 |
0.75 |
6.771649075 |
-37.85357323 |
75.98973334 |
0.389941108 |
0.9 |
2.36E-04 |
-0.178058059 |
-0.048797293 |
0.129260766 |
0.75 |
7.182176292 |
-40.14842369 |
61.76636997 |
0.34908304 |
Ml of ligand |
[L] X10-4 |
(Ep,a)M |
(Ep,a)C |
∆E mv |
j |
Log βj |
∆G (KJ/mol) |
∆H (KJ/mol) |
∆S (KJ/mol) |
0.9 |
2.36E-04 |
0.1955915 |
0.326947691 |
0.131356191 |
0.75 |
7.254511691 |
-40.55277916 |
61.15049185 |
0.348358524 |
0.9 |
2.36E-04 |
0.213753827 |
0.327533031 |
0.113779204 |
0.75 |
6.647742842 |
-37.16093638 |
66.73196731 |
0.35585855 |
0.9 |
2.36E-04 |
0.186240013 |
0.294298648 |
0.108058635 |
0.75 |
6.450265137 |
-36.05703441 |
68.77499585 |
0.359075288 |
0.9 |
2.36E-04 |
0.185063824 |
0.308701313 |
0.123637489 |
0.75 |
6.988057154 |
-39.06329614 |
63.48215938 |
0.351243211 |
On comparing the different thermodynamic parameters .mainly the stability constants and Gibbs free energies of complex interaction [9-32] for the reaction of oxalic acid plus copper bromide by analysis of the two couples of peaks at different san rates, we noticed almost equal values were obtained by analysis of the first and second couples of peaks as the data represented in Tables 11 & 12.
3.3. Quantum Chemical Calculations
The different quantum chemical calculation were done using Gaussian 09 program and set DFT B3LYP/6-311G method of calculation [29]. From the many obtained data we obtained the HOMO and LUMO energy levels for oxalic acid. The difference between the two mentioned energy levels gave the gap energy which is good and indicate its reactivity Appendix 1.
| Thermal property | Oxalic acid |
Oxalic acid-CuBr 2 complex |
| Dipole moment | 0 |
4.9107 Debye |
| Quadrupole moment (XX) | -32.2683 |
-71.654 |
| Octapole moment (XXX) | 0 |
0.204 |
| Hexadecapole moment (XXXX) | -36.7838 |
-19.1175 |
| Translational energy | 0.889 k.cal/Mol-K |
0.889 k.cal?Mol-K |
| Heat capacity at constant volume (Cv) | 2.981 cal/Mol.K |
2.981 cal/Mol.K |
| Translational entropy | 39.404 cal/Mol.K |
42.314 cal/Mol.K |
3.4. Comparison between Oxalic Acid and Oxalic Acid –Cubr2 Complex in Thermal Properties,
The different thermal properties were evaluated from the theoretical calculation of the frequencies for both oxalic acid alone and oxalic acid-CuBr2 complex quantum mechanically in water .The main data is summarized in Table 13 and the whole data are given in Appendixes 2 and 3. It was observed that the complex oxalic acid-CuBr2 has greater dipole moment than that of oxalic acid alone indicate its activity. The quadrupole moment. Octapole moment, heat capacity at constant volume and translational entropy are greater for the complex oxalic acid –copper bromide ,the we can use as active agent in biological application. All the complex parameter facilitate its uses .Other parameters are given in Appendixes 2 and 3.
Absolute solvation free energy of hydrogen ion under complexing conditions of CuBr2 + Oxalic acid:
Estimation of real solvation free energy of the proton and the value of ESHE which determined by the cycle [30].:
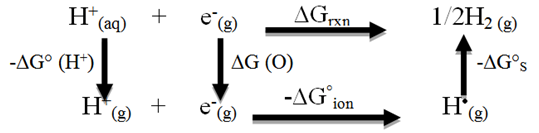
When we stdy the cycle given above we can estimate the ESHE the standard hydrogen electrode potential under our conditions applying equation ( 3 )and the ΔG°ion under the complex reaction between CuBr2 and oxalic acid.
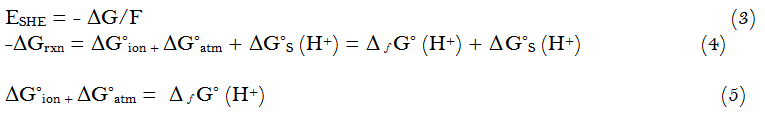
The coresponding Δ f G° (H+) based on Fermi-Dirack statistics is 1095 k.J/mole [31, 32].
Applying the above cycle the solvation free energy for hydrogen ion ΔG°S (H+) can be calculated applying equation (4) for our results and gave values ranging from -1131kJ/Mol at scan rate 0.05 V/Sec to -1135 kJ/Mol at scan rate 0.01 V/Sec for interaction of CuBr2 with oxalic acid in 0.1M KBr.This indicate that the san rate has little effect of the solvation of hydrogen ion.The sum of ΔG°ion + ΔG°atm for hydrogen ion was evaluated by applying equation (5) and know that ΔG°atm is concluded in all mmeasurents ,therefore the values of ΔG°ion + ΔG°atm indication about the ionic solvation free energy of hydrogen ion with values are ranging from 1056 KJ/Mol at 0.1M scan rate to 1135 KJ/Mol at scan rate 0.01 V/Sec.This indicate the complex formed from the interaction of CuBr2 and oxalic acid decrease the hydroen ion activity.The free energy of hydrogen ion is decreased when the complex is formed between the reaction of CuBr2 and oxalic acid indicating the hydrogen ion is less movable than in medium without complex interaction.
It was also observed that many theoretical data for the complex studies are greater than that of oxalic acid alone. As example all the partition function data are greater for the complex than the used ligand specially the electronic and rotational partition functions which gave corresponding ally greater energy values.
4. Conclusions
CuBr2 peaks are explained in absence and presence of oxalic acid as a ligand. Different solvation parameters were evaluated by analysis of the two couples of peaks for copper ions. The thermodynamic complexation properties were evaluated and found insure the complexation interaction between cupper bromide and oxalic acid studied here.
References
[1] T. Libuse, L. Zerzankova, F. Dycka, R. Mikelova, and F. Jelen, "Study of copper and purine-copper complexes on modified carbon electrodes by cyclic and elimination voltammetry," Sensors, vol. 8, pp. 429-444, 2008. Available at: https://doi.org/10.3390/s8010429.
[2] P. T. Kissinger and W. R. Heineman, "Cyclic voltammetry," Journal Chem Education, vol. 60, pp. 702-706, 1983.
[3] S. Rodrigues, A. Shukla, and N. Munichandraiah, "A cyclic voltammetric study of the kinetics andmechanism of electrodeposition of manganese dioxide," Journal of Applied Electrochemistry, vol. 28, pp. 1235-1241, 1998.
[4] C. M. A. Brett and A. M. O. Brett, Electrochemistry principles, methods and application, 1993 ed. Oxford: Oxford University Press, 1993.
[5] A. Khan, R. Ahmed, and M. Mirza, "Evaluation of kinetic parameters of uranyl acetate complexes in ethanolic solution by cyclic voltammetry," Journal of Radioanalytical and Nuclear Chemistry, vol. 283, pp. 527-531, 2010. Available at: https://doi.org/10.1007/s10967-009-0372-4.
[6] H. Matsuda and Y. Ayabe, "The theory of the cathode-ray polarography of randles-sevcik," Zeitschrift fuer Elektrochmie and Angewandte Physikalische Chemie, vol. 59, pp. 494-503, 1955.
[7] D. K. Grosser, Cyclic voltammetry: Simulation and analysis of reaction mechanisms vol. 43. New York: VCH, 1993.
[8] C. H. Bamford, C. F. Tipper, H., and R. Compton, G., Electrode Kinetics: Principles and methology vol. 26. New York: Elsevier, 1986.
[9] D. A. C. Brownson and C. E. Banks, The handbook of graphene electrochemistry. London: Springer-Verlag, 2014.
[10] Y. Wang, R. M. Hernandez, D. J. Bartlett, J. M. Bingham, T. R. Kline, A. Sen, and T. E. Mallouk, "Bipolar electrochemical mechanism for the propulsion of catalytic nanomotors in hydrogen peroxide solutions," Langmuir, vol. 22, pp. 10451-10456, 2006. Available at: https://doi.org/10.1021/la0615950.
[11] A. M. E. H. El-Askalany and A. M. Abou El-Magd, "Stability constants of Zn(II),Pd(II),Cd(II) and Cu(II) complexes with hematoxylin," Chemical and Pharmaceutical Bulletin, vol. 43, pp. 1791-1792, 1995.
[12] E. A. Gomaa and R. M. Abu-Qarn, "Ionic association and thermodynamic parameters for solvation of vanadyl sulfate in ethanol-water mixtures at different temperatures," Journal of Molecular Liquids, vol. 232, pp. 319-324, 2017. Available at: https://doi.org/10.1016/j.molliq.2017.02.085.
[13] E. A. Gomaa and M. A. Tahoon, "Ion association and solvation behavior of copper sulfate in binary aqueous–methanol mixtures at different temperatures," Journal of Molecular Liquids, vol. 214, pp. 19-23, 2016. Available at: https://doi.org/10.1016/j.molliq.2015.11.046.
[14] E. Gomaa, R. Zaky, and A. Shokr, "Estimated the physical parameters of lanthanum chloride in water-N, N-dimethyl formamide mixtures using different techniques," Journal of Molecular Liquids, vol. 242, pp. 913-918, 2017. Available at: https://doi.org/10.1016/j.molliq.2017.07.108.
[15] E. Gomaa, R. Zaky, and A. Shokr, "Effect of calcon carboxylic acid on association process of vanadyl sulfate in water-N, N-dimethyl formamide mixed solvents," Chemical Data Collections, vol. 11, pp. 67-76, 2017. Available at: https://doi.org/10.1016/j.cdc.2017.08.002.
[16] E. A. Gomaa, A. Negm, and M. A. Tahoon, "Conductometric and volumetric study of copper sulphate in aqueous ethanol solutions at different temperatures," Journal of Taibah University for Science, vol. 11, pp. 741-748, 2017. Available at: https://doi.org/10.1016/j.jtusci.2016.08.007.
[17] S. El-Shereafy, E. Gomaa, A. Yousif, and A. Abou Elyazed, "Electrochemical and thermodynamic estimations of the interaction parameters for bulk and nano-silver nitrate (NSN) with cefdinir drug using a glassy carbon electrode," Iranian Journal of Materials Science and Engineering, vol. 14, pp. 48-57, 2017.
[18] J. I. Kim, A. Cecal, H. J. Born, and E. A. Gomaa, "Preferential solvation of single Ion: Acritical study of the Ph4AsPh4B assumption for single ion thermodynamics in mixed aqueous-acetonitrile and aqueous-N,N-dimethyl formamide solvents, Z," Physical Chemistry, vol. 110, pp. 209-227, 1978.
[19] J. Kim. and E. Gomaa, "Preferential solvation of single Ions: The PH4ASPH4B assumption for single Ion thermodynamics in mixed dimethylsulfoxide-water dolvents," Bulletin des Sociétés Chimiques Belges, vol. 90, pp. 391-407, 1981. Available at: https://doi.org/10.1002/bscb.19810900415.
[20] M. Ghandour, R. Abo-Doma, and E. Gomaa, "The electroreduction (polarographically) of uranyl ion in nitric acid and nitric acid-methanol mixture media," Electrochimica Acta, vol. 27, pp. 159-163, 1982. Available at: https://doi.org/10.1016/0013-4686(82)80075-3.
[21] E. Gomaa, "Thermodynamic studies of the solvation of Ph4AsPh4B in mixed solvents (MeOH—DMF)," Thermochimica Acta, vol. 80, pp. 355-359, 1984. Available at: https://doi.org/10.1016/0040-6031(84)87214-7.
[22] E. A. Gomaa, A. E. Negm, and R. M. Abu Qarn, "Cyclic voltammetry of lead nitrate with acetyl acetone using glassy carbon electrode," AASCIT, vol. 3, pp. 71-76, 2016.
[23] A. M. El-Hady, E. A. Gomaa, and A. G. Al-Harazie, "Cyclic voltammetry of bulk and nano CdCl2 with ceftazidime drug and some DFT calculations," Journal of Molecular Liquids, vol. 276, pp. 970-985, 2019.
[24] R. S. Nicholson and I. Shain, "Theory and application of cyclic voltammetry for measurement of electrode reaction kinetics," Analytical Chemistry, vol. 37, pp. 178-190, 1965.
[25] G. A. Mabbott, "An introduction to cyclic voltammetry," Journal of Chemical Education, vol. 60, pp. 697-702, 1983.
[26] E.-H. M. Abd, E. Gomaa, R. Zaky, and A. Gomaa, "Synthesis, characterization, computational simulation, cyclic voltammetry and biological studies on Cu (II), Hg (II) and Mn (II) complexes of 3-(3, 5-dimethylpyrazol-1-yl)-3-oxopropionitrile," Journal of Molecular Liquids, p. 112794, 2020.
[27] E. A. Gomaa, M. H. Mahmoud, M. G. Mousa, and E. M. El-Dahshan, "Cyclic voltammetry for the interaction between bismuth nitrate and methyl red in potassium nitrate solutions," Chemical Methodologies, vol. 3, pp. 1-11, 2018.
[28] A. Brolo, M. Temperini, and S. Agostinho, "Copper dissolution in bromide medium in the absence and presence of hexamethylenetetramine (HMTA)," Electrochimica Acta, vol. 44, pp. 559-571, 1998. Available at: https://doi.org/10.1016/s0013-4686(98)00179-0.
[29] E. A. Gomaa and R. T. Rashad, "Thermal and thermodynamic parameters for glycine (GL) solvation in water theoretically," Biomedical Journal of Scientific & Technical Research, vol. 23, pp. 17345-17348, 2019.
[30] K. P. Casey, C. J. Cramer, and D. G. Truhlar, "Aqueous solvation free energies of ions and ion− water clusters based on an accurate value for the absolute aqueous solvation free energy of the proton," The Journal of Physical Chemistry B, vol. 110, pp. 16066-16081, 2006. Available at: https://doi.org/10.1021/jp063552y.
[31] P. Casey, C. J. Kelly, and D. G. Cramer, "Truhlar, single ion solvation free energies and the normal hydrogen electrode in methanol, acetonitrile, dimethylsulfoxide," Journal Physics Chem B, vol. 2, pp. 408-422, 2007.
[32] W. Paul, C. J. Cramer, and D. G. Truhlar, "Computation of equilibrium oxidation and reduction potentials for reversible and dissociative electron-transfer reactions in solution," Theoretical Chemistry Accounts, vol. 112, pp. 217-227, 2004. Available at: https://doi.org/10.1007/s00214-004-0577-0.
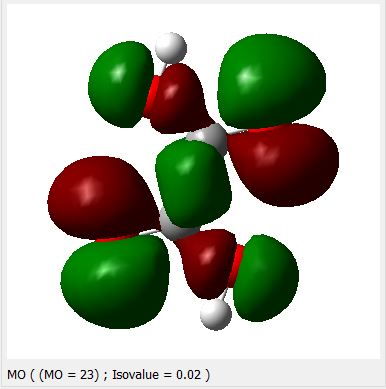
HOMO, (N=23)(-0.30111ev)
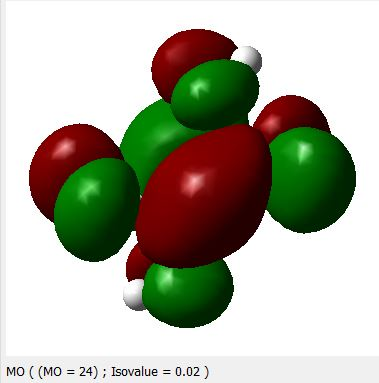
Appendix-1. HOMO & LUMO levels for oxalic acid.
Appendix-2. Physical properties of oxalic acid alone.
Dipole moment (field-independent basis, Debye):
X= 0.0000 Y= 0.0000 Z= 0.0000 Tot= 0.0000
Quadrupole moment (field-independent basis, Debye-Ang):
XX= -45.9262 YY= -19.1403 ZZ= -32.2683
XY= 1.3844 XZ= 0.0000 YZ= 0.0000
Traceless Quadrupole moment (field-independent basis, Debye-Ang):
XX= -13.4813 YY= 13.3046 ZZ= 0.1766
XY= 1.3844 XZ= 0.0000 YZ= 0.0000
Octapole moment (field-independent basis, Debye-Ang**2):
XXX= 0.0000 YYY= 0.0000 ZZZ= 0.0000 XYY= 0.0000
XXY= 0.0000 XXZ= 0.0000 XZZ= 0.0000 YZZ= 0.0000
YYZ= 0.0000 XYZ= 0.0000
Hexadecapole moment (field-independent basis, Debye-Ang**3):
XXXX= -268.2226 YYYY= -167.5877 ZZZZ= -27.9062 XXXY= -36.7838
XXXZ= 0.0000 YYYX= -6.8845 YYYZ= 0.0000 ZZZX= 0.0000
ZZZY= 0.0000 XXYY= -102.5791 XXZZ= -43.7167 YYZZ= -58.0004
XXYZ= 0.0000 YYXZ= 0.0000 ZZXY= -6.9493
N-N= 2.311045083462D+02 E-N=-1.350788684135D+03 KE= 3.749716196477D+02
Symmetry AG KE= 1.811837398941D+02
Symmetry BG KE= 8.499432461333D+00
Symmetry AU KE= 8.032598844166D+00
Symmetry BU KE= 1.772558484481D+02
Exact polarizability: 60.502 -2.061 50.158 0.000 0.000 27.232
Zero-point correction= 0.048716 (Hartree/Particle)
Thermal correction to Energy= 0.054282
Thermal correction to Enthalpy= 0.055226
Thermal correction to Gibbs Free Energy= 0.019237
Sum of electronic and zero-point Energies= -378.311469
Sum of electronic and thermal Energies= -378.305903
Sum of electronic and thermal Enthalpies= -378.304959
Sum of electronic and thermal Free Energies= -378.340948
E (Thermal) CV S
KCal/Mol Cal/Mol-Kelvin Cal/Mol-Kelvin
Total 34.062 18.538 75.744
Electronic 0.000 0.000 0.000
Translational 0.889 2.981 39.404
Rotational 0.889 2.981 24.923
Vibrational 32.285 12.576 11.417
Vibration 1 0.594 1.984 5.803
Vibration 2 0.675 1.727 1.590
Vibration 3 0.789 1.410 0.866
Vibration 4 0.794 1.399 0.850
Vibration 5 0.885 1.184 0.587
Vibration 6 0.977 0.995 0.421
Q Log10(Q) Ln(Q)
Total Bot 0.141765D-08 -8.848430 -20.374263
Total V=0 0.362433D+14 13.559228 31.221276
Vib (Bot) 0.676668D-21 -21.169625 -48.744862
Vib (Bot) 1 0.681126D+01 0.833228 1.918578
Vib (Bot) 2 0.712891D+00 -0.146977 -0.338426
Vib (Bot) 3 0.407968D+00 -0.389373 -0.896565
Vib (Bot) 4 0.401807D+00 -0.395983 -0.911784
Vib (Bot) 5 0.301784D+00 -0.520303 -1.198043
Vib (Bot) 6 0.237511D+00 -0.624316 -1.437541
Vib (V=0) 0.172995D+02 1.238033 2.850677
Vib (V=0) 1 0.732959D+01 0.865080 1.991920
Vib (V=0) 2 0.137075D+01 0.136960 0.315362
Vib (V=0) 3 0.114532D+01 0.058927 0.135684
Vib (V=0) 4 0.114144D+01 0.057454 0.132293
Vib (V=0) 5 0.108402D+01 0.035035 0.080672
Vib (V=0) 6 0.105354D+01 0.022653 0.052160
Electronic 0.100000D+01 0.000000 0.000000
Appendix-3. Cu oxalate theoretical data.
Dipole moment (field-independent basis, Debye):
X= 0.0663 Y= 4.9102 Z= 0.0000 Tot= 4.9107
Quadrupole moment (field-independent basis, Debye-Ang):
XX= -91.9474 YY= -114.3172 ZZ= -71.6546
XY= -0.2874 XZ= 0.0000 YZ= 0.0000
Traceless Quadrupole moment (field-independent basis, Debye-Ang):
XX= 0.6923 YY= -21.6775 ZZ= 20.9852
XY= -0.2874 XZ= 0.0000 YZ= 0.0000
Octapole moment (field-independent basis, Debye-Ang**2):
XXX= 0.8198 YYY= 55.7168 ZZZ= 0.0000 XYY= 0.2044
XXY= 20.5202 XXZ= 0.0000 XZZ= 0.0291 YZZ= 1.9610
YYZ= 0.0000 XYZ= 0.0000
Hexadecapole moment (field-independent basis, Debye-Ang**3):
XXXX= -705.9591 YYYY= -3600.9243 ZZZZ= -59.2390 XXXY= -19.1175
XXXZ= 0.0000 YYYX= -18.5880 YYYZ= 0.0000 ZZZX= 0.0000
ZZZY= 0.0000 XXYY= -724.5780 XXZZ= -118.5522 YYZZ= -458.3488
Zero-point correction= 0.051032 (Hartree/Particle)
Thermal correction to Energy= 0.063515
Thermal correction to Enthalpy= 0.064459
Thermal correction to Gibbs Free Energy= 0.009653
Sum of electronic and zero-point Energies= -2394.538381
Sum of electronic and thermal Energies= -2394.525898
Sum of electronic and thermal Enthalpies= -2394.524954
Sum of electronic and thermal Free Energies= -2394.579760
E (Thermal) CV S
KCal/Mol Cal/Mol-Kelvin Cal/Mol-Kelvin
Total 39.856 42.355 115.348
Electronic 0.000 0.000 1.377
Translational 0.889 2.981 42.314
Rotational 0.889 2.981 31.566
Vibrational 38.079 36.393 40.090
Vibration 1 0.594 1.983 5.556
Vibration 2 0.594 1.981 5.210
Vibration 3 0.602 1.957 3.672
Vibration 4 0.604 1.949 3.463
Vibration 5 0.606 1.942 3.281
Vibration 6 0.628 1.870 2.358
Vibration 7 0.643 1.825 2.039
Vibration 8 0.658 1.778 1.796
Vibration 9 0.662 1.766 1.744
Vibration 10 0.691 1.677 1.428
Vibration 11 0.704 1.641 1.324
Vibration 12 0.725 1.581 1.180
Vibration 13 0.739 1.542 1.097
Vibration 14 0.788 1.414 0.872
Vibration 15 0.791 1.406 0.861
Vibration 16 0.806 1.367 0.804
Vibration 17 0.825 1.321 0.744
Vibration 18 0.945 1.058 0.471
Vibration 19 0.954 1.039 0.455
Q Log10(Q) Ln(Q)
Total Bot 0.362864D-04 -4.440256 -10.224068
Total V=0 0.107895D+20 19.032999 43.825101
Vib (Bot) 0.707643D-19 -19.150186 -44.094933
Vib (Bot) 1 0.601198D+01 0.779018 1.793755
Vib (Bot) 2 0.504664D+01 0.703002 1.618722
Vib (Bot) 3 0.229876D+01 0.361493 0.832369
Vib (Bot) 4 0.206136D+01 0.314153 0.723365
Vib (Bot) 5 0.187405D+01 0.272781 0.628101
Vib (Bot) 6 0.113443D+01 0.054777 0.126128
Vib (Bot) 7 0.943237D+00 -0.025379 -0.058437
Vib (Bot) 8 0.813282D+00 -0.089759 -0.206678
Vib (Bot) 9 0.787101D+00 -0.103969 -0.239399
Vib (Bot) 10 0.638740D+00 -0.194676 -0.448258
Vib (Bot) 11 0.593359D+00 -0.226682 -0.521956
Vib (Bot) 12 0.532801D+00 -0.273435 -0.629607
Vib (Bot) 13 0.498781D+00 -0.302090 -0.695587
Vib (Bot) 14 0.410447D+00 -0.386743 -0.890508
Vib (Bot) 15 0.405957D+00 -0.391520 -0.901509
Vib (Bot) 16 0.384208D+00 -0.415433 -0.956571
Vib (Bot) 17 0.361154D+00 -0.442307 -1.018450
Vib (Bot) 18 0.257202D+00 -0.589726 -1.357894
Vib (Bot) 19 0.251126D+00 -0.600108 -1.381800
Vib (V=0) 0.210412D+05 4.323070 9.954236
Vib (V=0) 1 0.653274D+01 0.815095 1.876826
Vib (V=0) 2 0.557134D+01 0.745960 1.717636
Vib (V=0) 3 0.285251D+01 0.455227 1.048198
Vib (V=0) 4 0.262113D+01 0.418489 0.963606
Vib (V=0) 5 0.243960D+01 0.387319 0.891835
Vib (V=0) 6 0.173973D+01 0.240481 0.553729
Vib (V=0) 7 0.156757D+01 0.195226 0.449524
Vib (V=0) 8 0.145469D+01 0.162770 0.374791
Vib (V=0) 9 0.143248D+01 0.156090 0.359411
Vib (V=0) 10 0.131117D+01 0.117657 0.270916
Vib (V=0) 11 0.127593D+01 0.105829 0.243679
Vib (V=0) 12 0.123067D+01 0.090141 0.207558
Vib (V=0) 13 0.120625D+01 0.081436 0.187513
Vib (V=0) 14 0.114689D+01 0.059522 0.137054
Vib (V=0) 15 0.114405D+01 0.058445 0.134575
Vib (V=0) 16 0.113057D+01 0.053297 0.122720
Vib (V=0) 17 0.111679D+01 0.047972 0.110460
Vib (V=0) 18 0.106227D+01 0.026237 0.060413
Vib (V=0) 19 0.105952D+01 0.025110 0.057817
Electronic 0.200000D+01 0.301030 0.693147
Translational 0.145127D+09 8.161749 18.793122
Rotational 0.176665D+07 6.247150 14.384595